CausalDynamics#
Here, we showcase some key features of CausalDynamics using small example graphs.
import torch
import matplotlib.pyplot as plt
from causaldynamics.scm import create_scm_graph
from causaldynamics.plot import plot_trajectories, plot_scm, plot_3d_trajectories
from causaldynamics.creator import create_scm, simulate_system
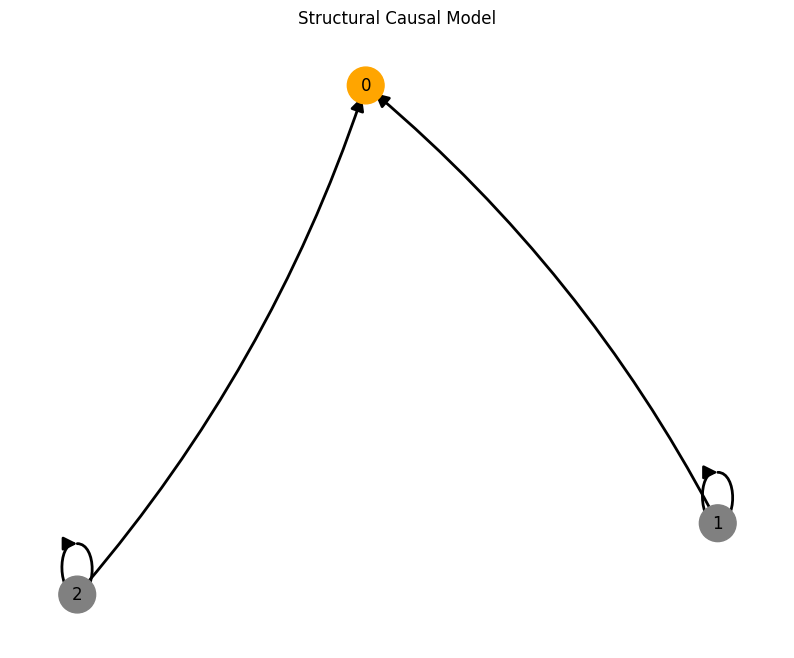
Basic coupled model#
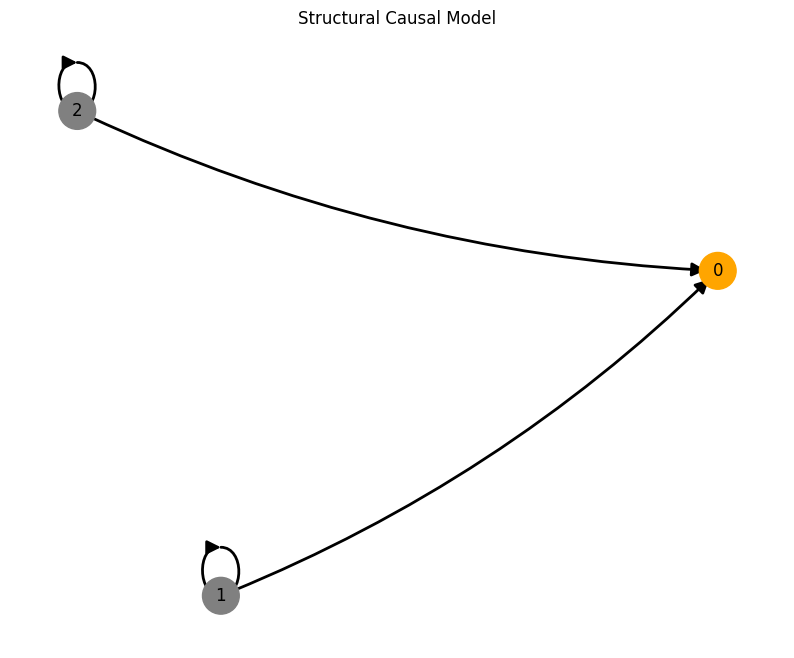
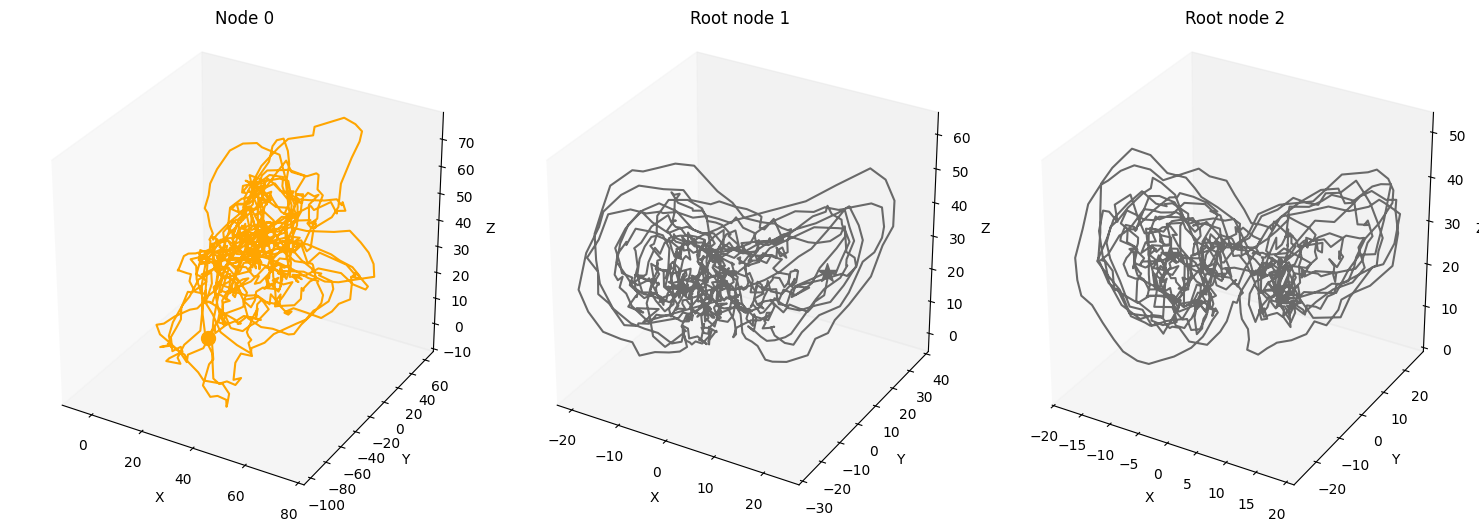
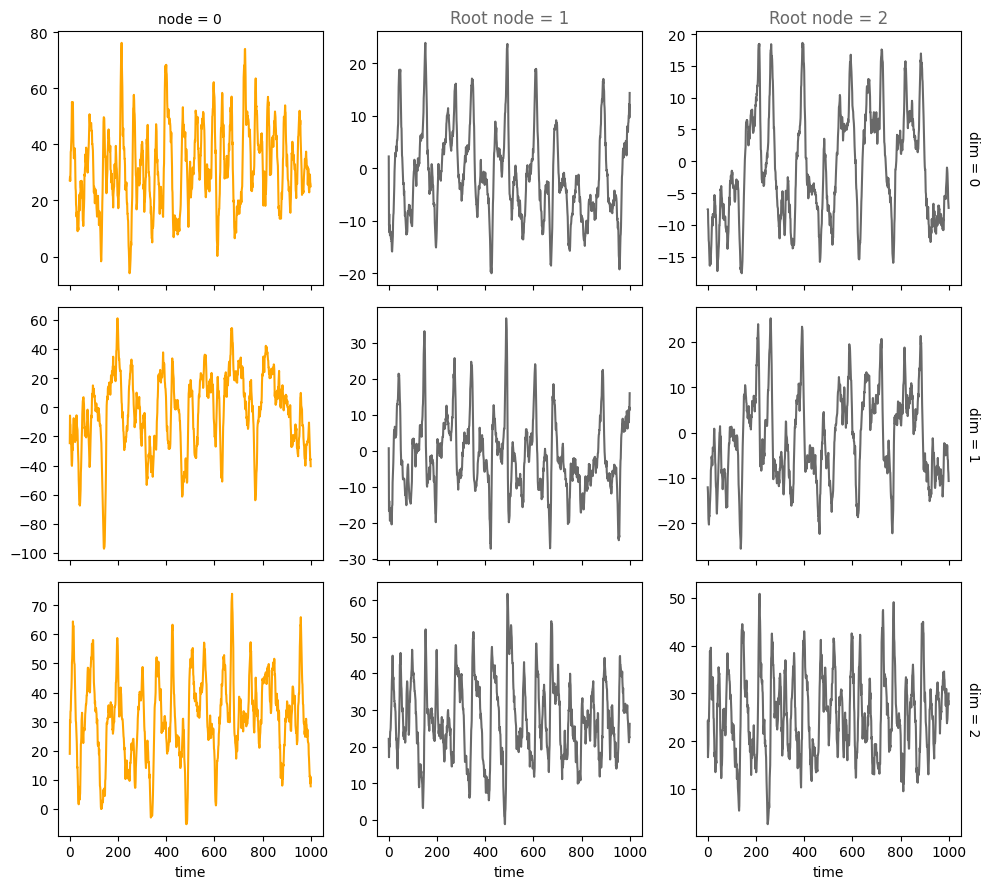
num_nodes = 3
node_dim = 3
num_timesteps = 1000
confounders=False,
system_name='Lorenz'
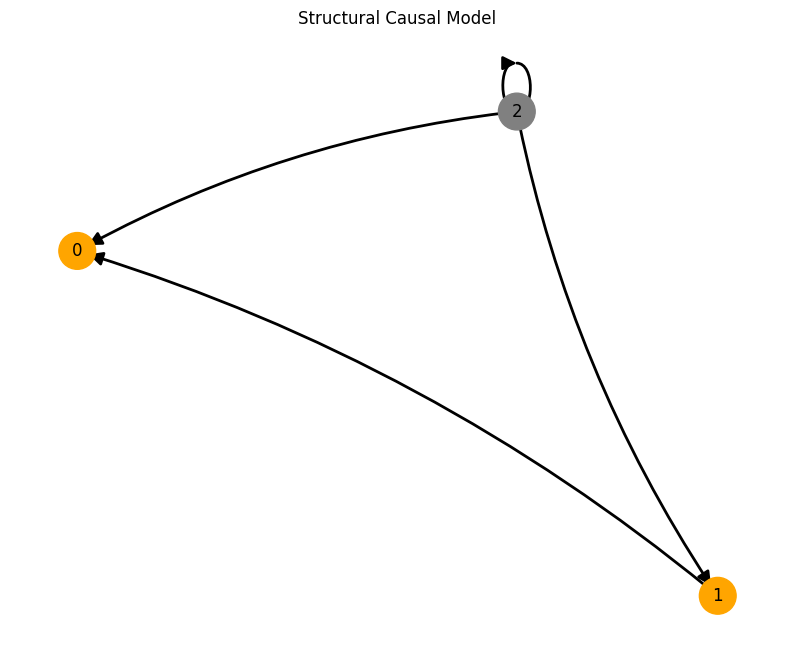
A = torch.tensor([[0,0,0],
[1,0,0],
[1,0,0]])
A, W, b, root_nodes, _ = create_scm(num_nodes,
node_dim=node_dim,
confounders=confounders,
adjacency_matrix=A)
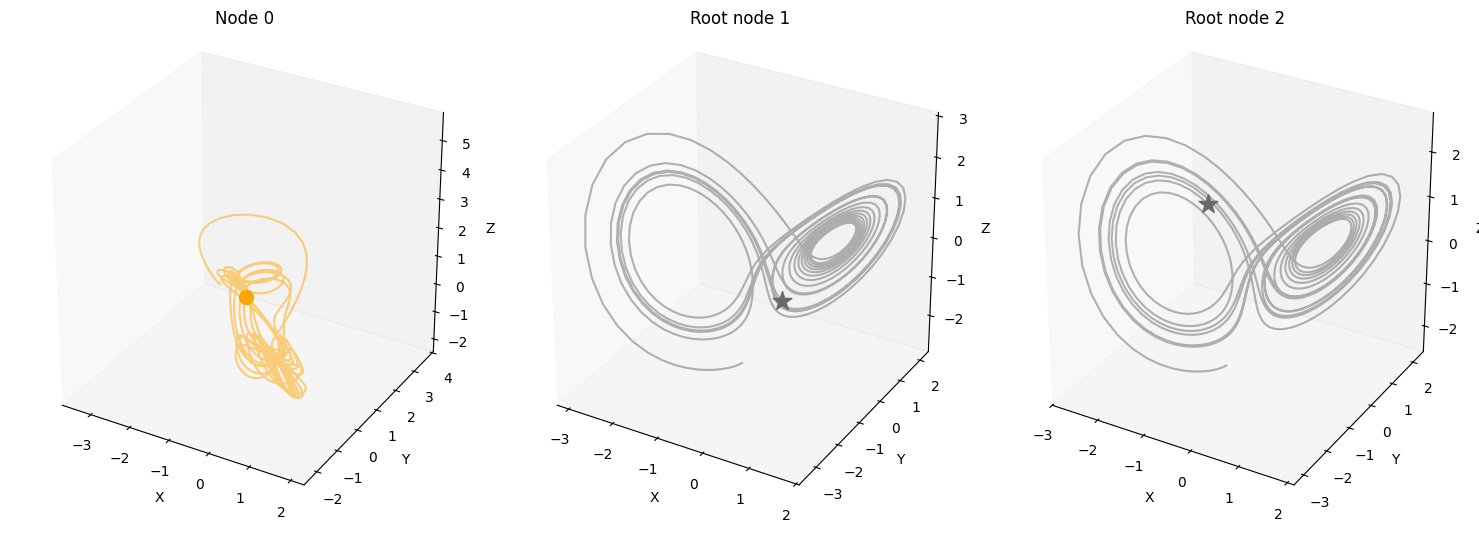
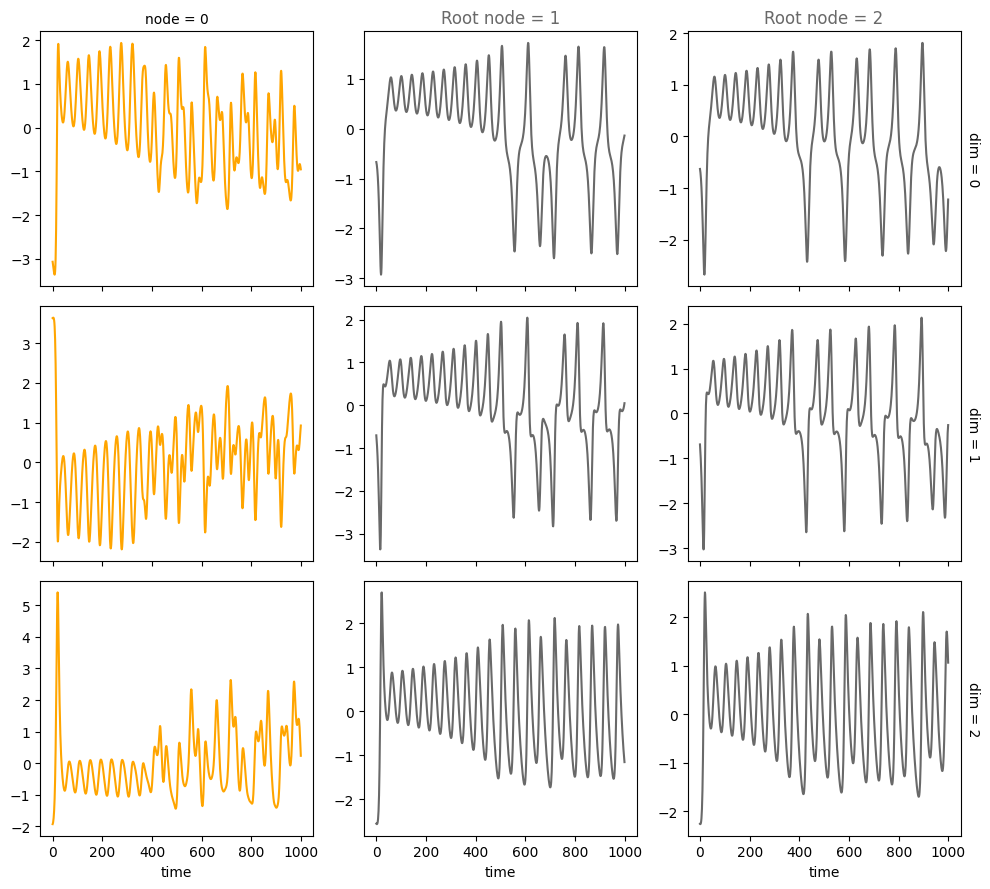
data = simulate_system(A, W, b,
num_timesteps=num_timesteps,
num_nodes=num_nodes,
system_name=system_name)
plot_scm(G=create_scm_graph(A), root_nodes=root_nodes)
plot_3d_trajectories(data, root_nodes, line_alpha=1.)
plot_trajectories(data, root_nodes, sharey=False)
plt.show()
INFO - Creating SCM with 3 nodes and 3 dimensions each...
INFO - Simulating Lorenz system for 1000 timesteps...
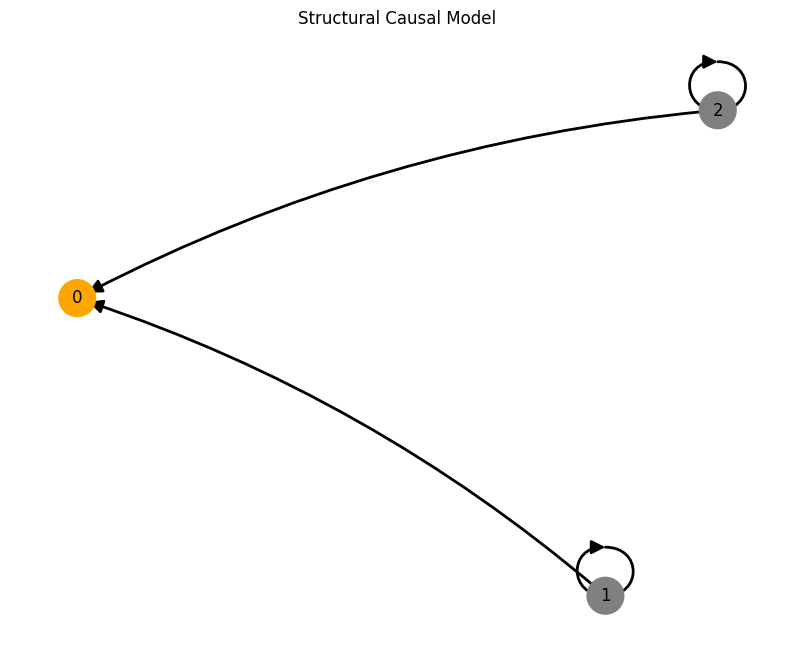
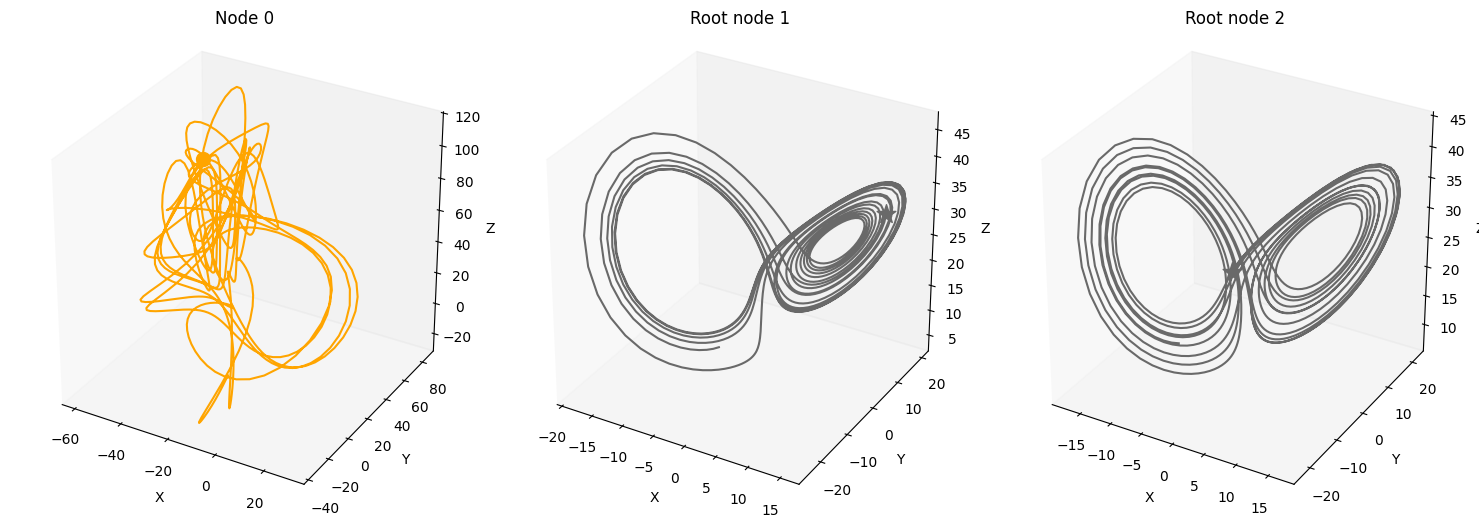
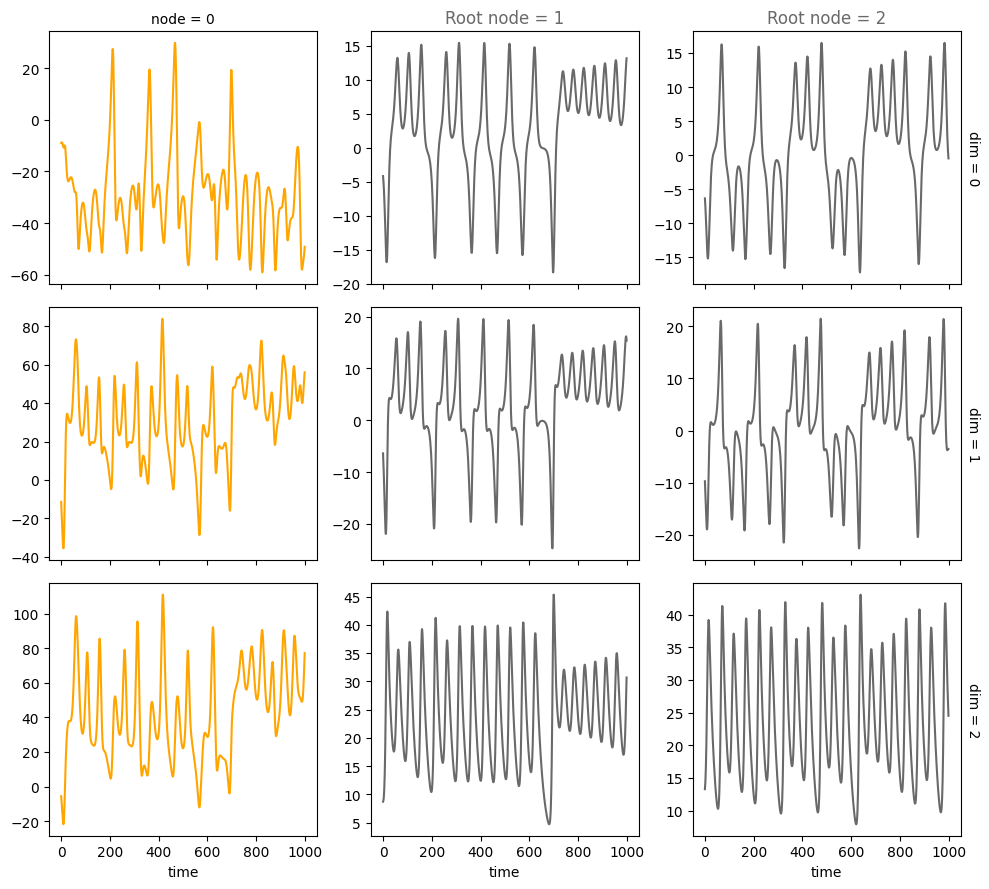
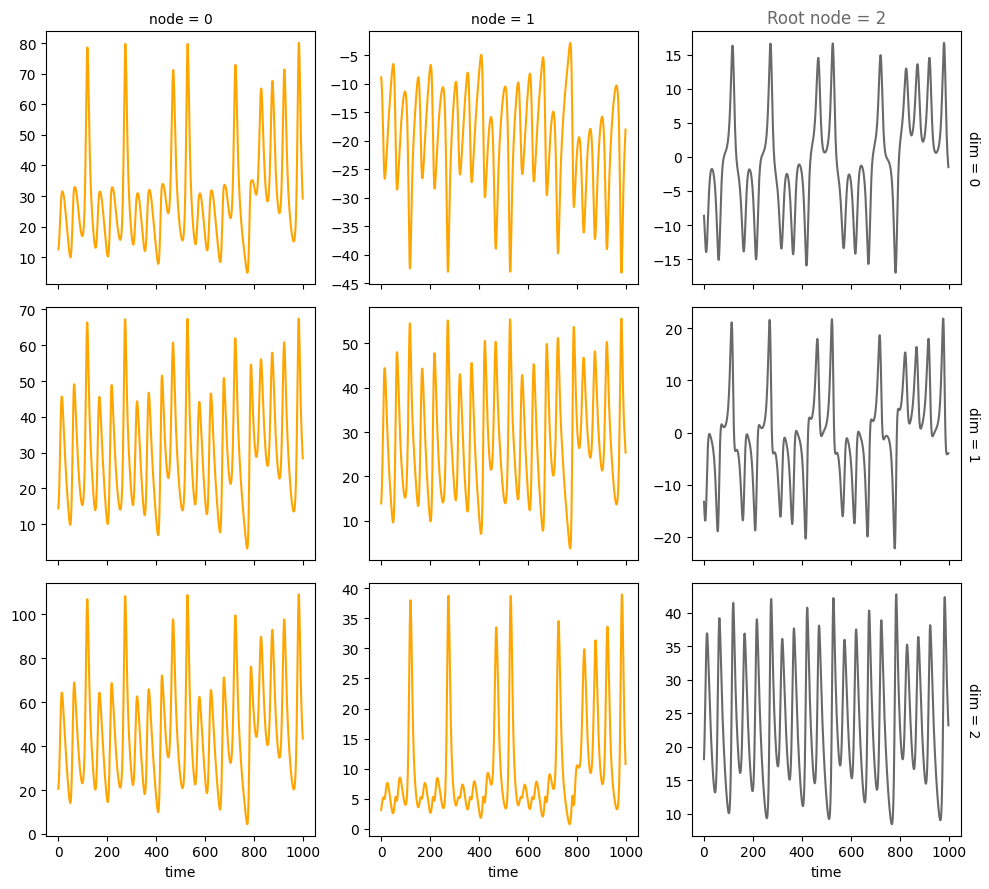
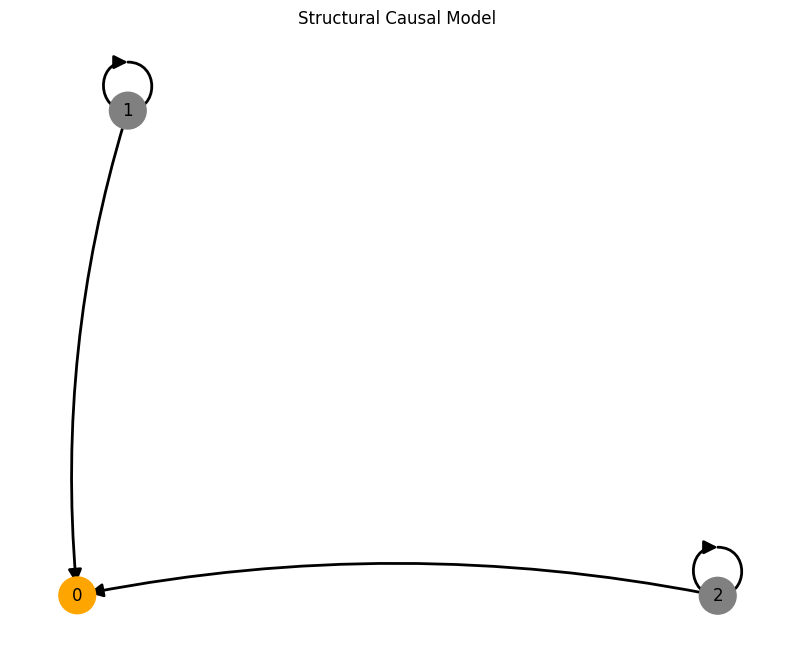
Confounders#
A = torch.tensor([[0,0,0],
[1,0,0],
[1,1,0]])
A, W, b, root_nodes, _ = create_scm(num_nodes,
node_dim=node_dim,
confounders=confounders,
adjacency_matrix=A)
data = simulate_system(A, W, b,
num_timesteps=num_timesteps,
num_nodes=num_nodes,
system_name=system_name)
plot_scm(G=create_scm_graph(A), root_nodes=root_nodes)
plot_3d_trajectories(data, root_nodes, line_alpha=1.)
plot_trajectories(data, root_nodes, sharey=False)
plt.show()
INFO - Creating SCM with 3 nodes and 3 dimensions each...
INFO - Simulating Lorenz system for 1000 timesteps...
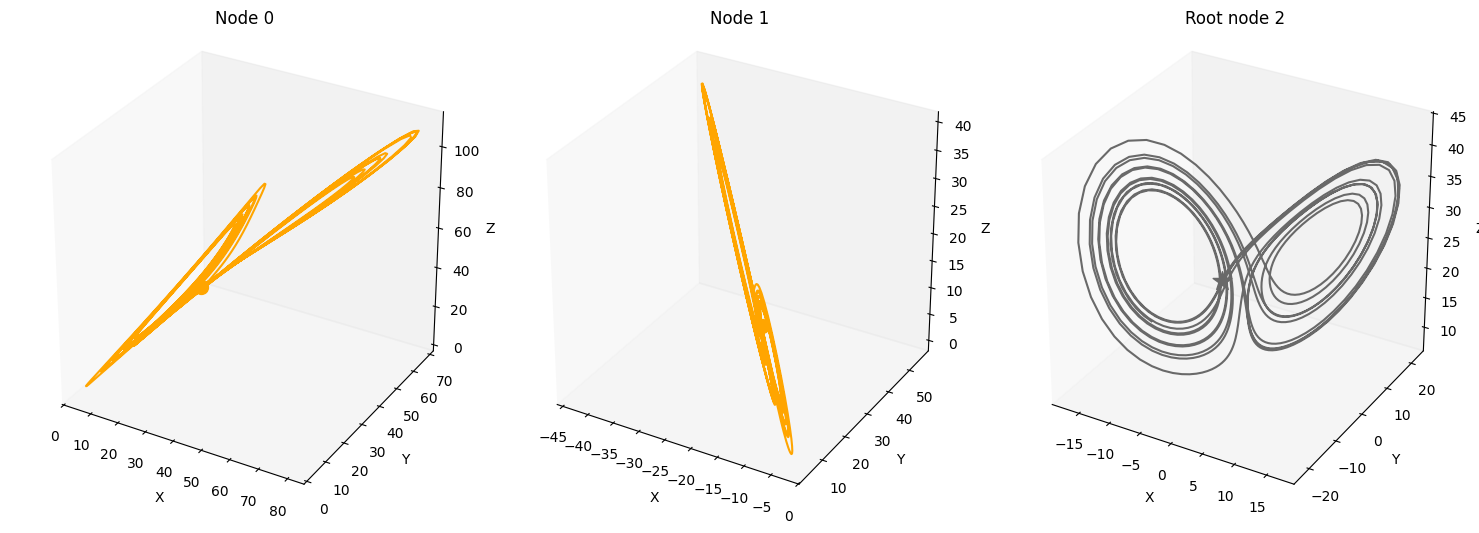
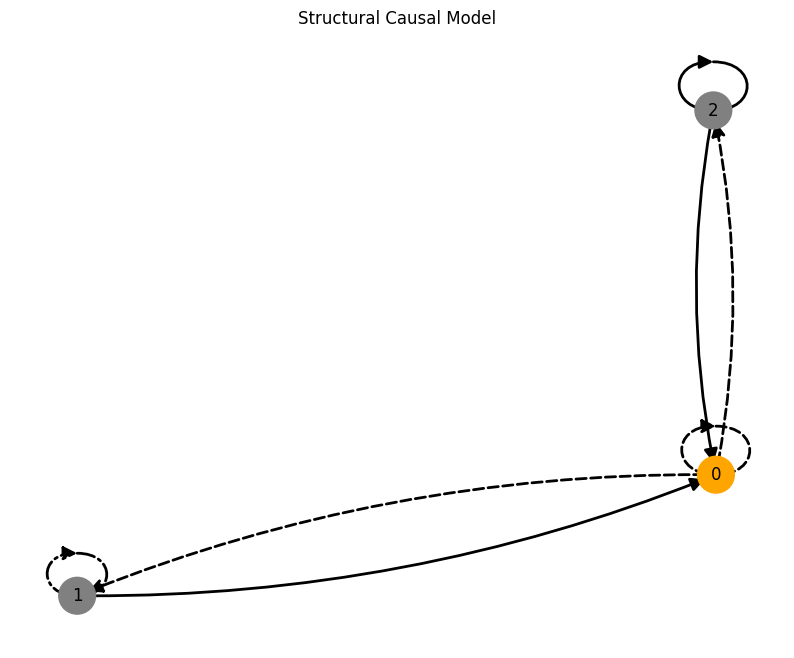
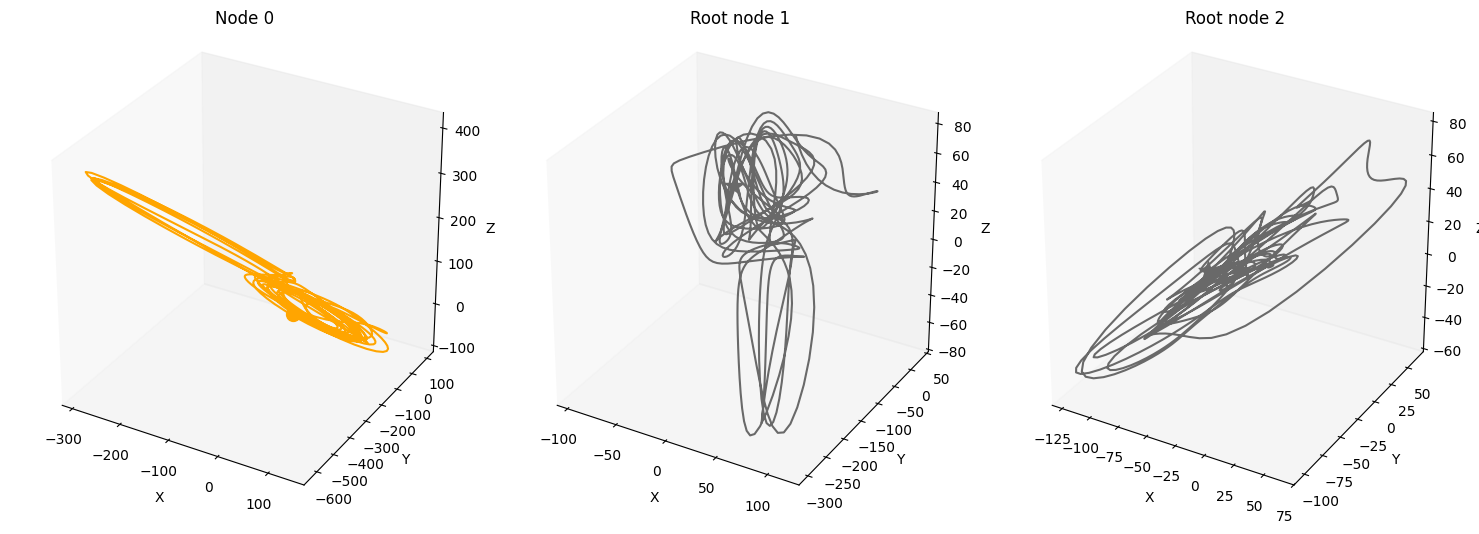
Time-lag edges#
time_lag = 10
time_lag_edge_probability = 0.1
A, W, b, root_nodes, magnitudes = create_scm(
num_nodes,
node_dim=node_dim,
confounders=False,
graph="scale-free",
time_lag=time_lag,
time_lag_edge_probability=time_lag_edge_probability
)
data = simulate_system(A, W, b,
num_timesteps=num_timesteps,
num_nodes=num_nodes,
system_name=system_name,
time_lag=time_lag,)
plot_scm(G=create_scm_graph(A), root_nodes=root_nodes)
plot_3d_trajectories(data, root_nodes, line_alpha=1.)
plot_trajectories(data, root_nodes, sharey=False)
plt.show()
INFO - Creating SCM with 3 nodes and 3 dimensions each...
INFO - Simulating Lorenz system for 1000 timesteps...
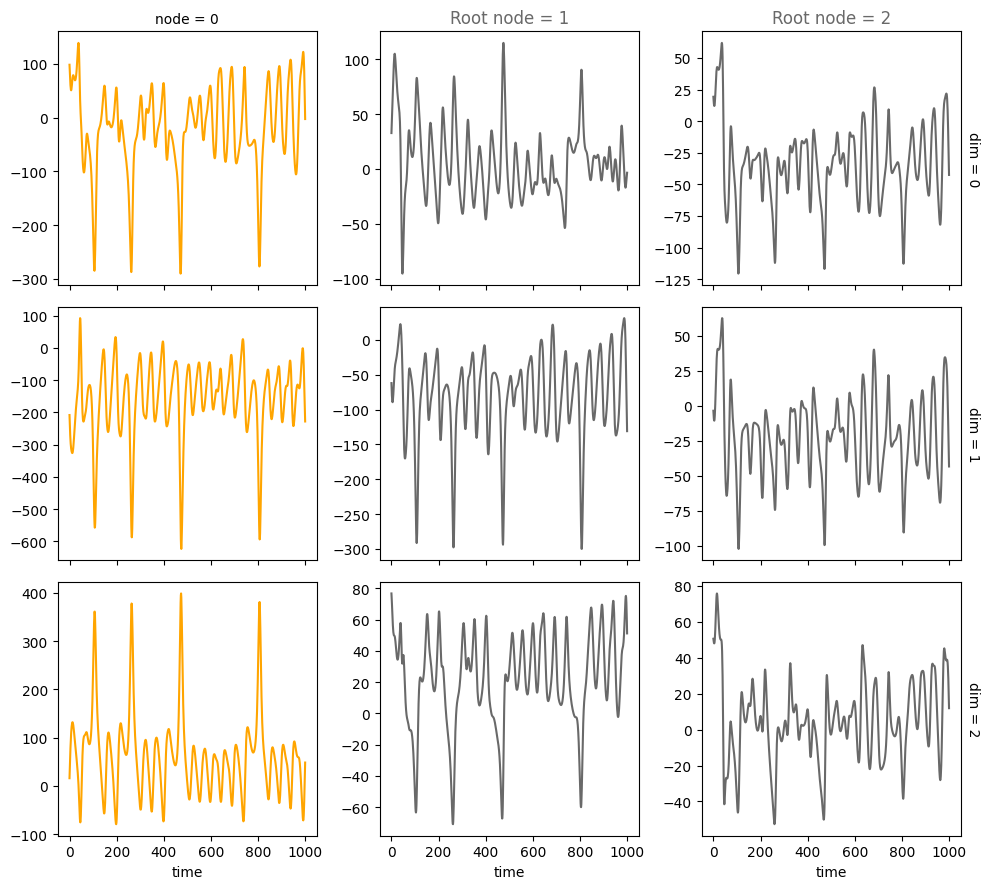
Stochastic systems#
num_nodes = 3
node_dim = 3
num_timesteps = 1000
noise = 1.
confounders=False,
system_name='Lorenz'
A = torch.tensor([[0,0,0],
[1,0,0],
[1,0,0]])
A, W, b, root_nodes, _ = create_scm(num_nodes,
node_dim=node_dim,
confounders=confounders,
adjacency_matrix=A)
data = simulate_system(A, W, b,
num_timesteps=num_timesteps,
num_nodes=num_nodes,
system_name=system_name,
make_trajectory_kwargs={'noise': noise})
plot_scm(G=create_scm_graph(A), root_nodes=root_nodes)
plot_3d_trajectories(data, root_nodes, line_alpha=1.)
plot_trajectories(data, root_nodes, sharey=False)
plt.show()
INFO - Creating SCM with 3 nodes and 3 dimensions each...
INFO - Simulating Lorenz system for 1000 timesteps...
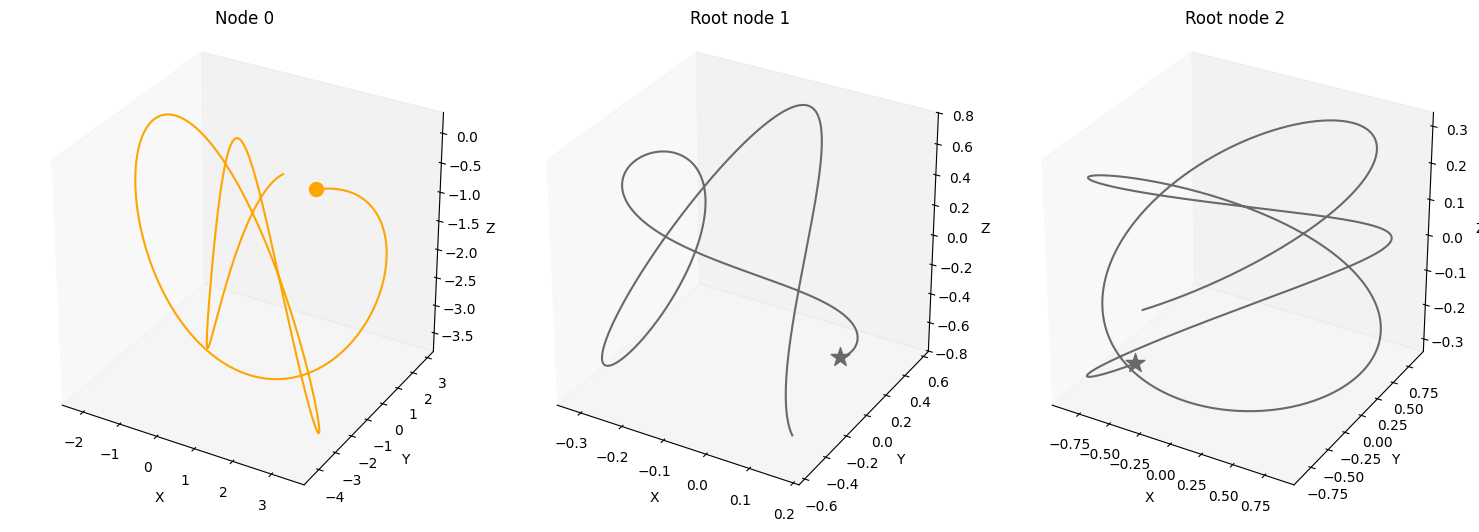
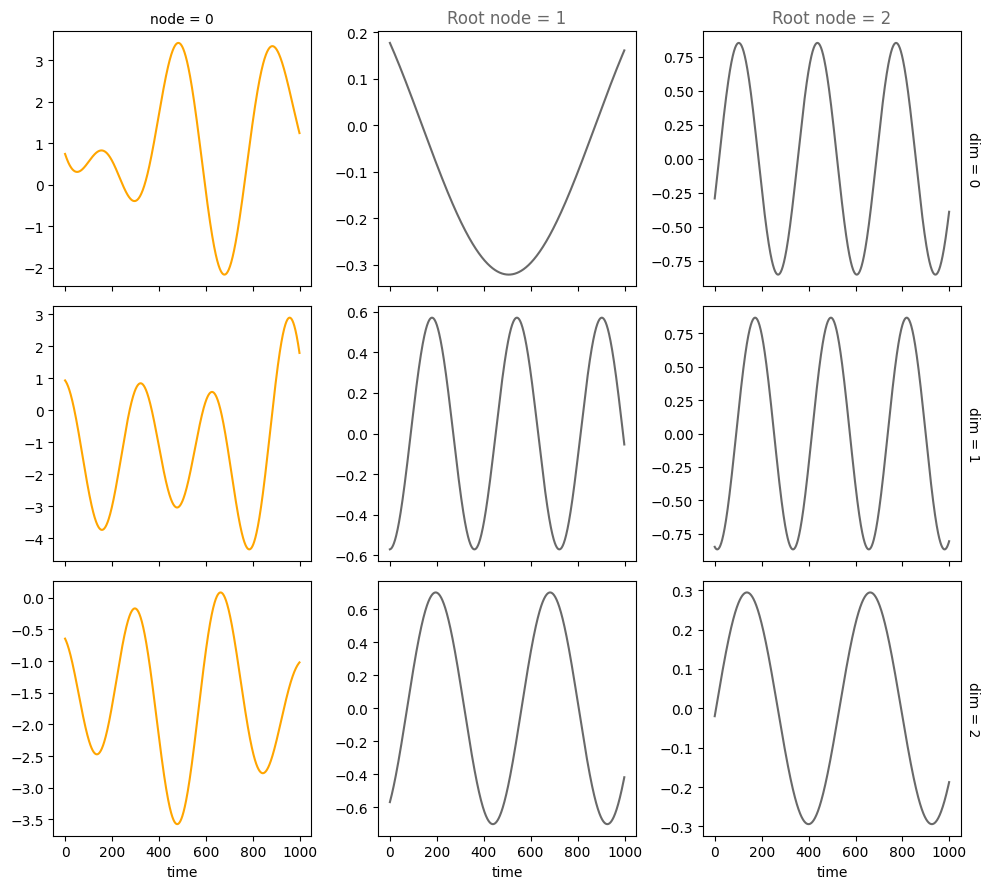
Periodic drivers#
num_nodes = 3
node_dim = 3
num_timesteps = 1000
confounders=False,
system_name='Lorenz'
A = torch.tensor([[0,0,0],
[1,0,0],
[1,0,0]])
A, W, b, root_nodes, _ = create_scm(num_nodes,
node_dim=node_dim,
confounders=confounders,
adjacency_matrix=A)
data = simulate_system(A, W, b,
num_timesteps=num_timesteps,
num_nodes=num_nodes,
system_name=system_name,
init_ratios=[1,1,0])
plot_scm(G=create_scm_graph(A), root_nodes=root_nodes)
plot_3d_trajectories(data, root_nodes, line_alpha=1.)
plot_trajectories(data, root_nodes, sharey=False)
plt.show()
INFO - Creating SCM with 3 nodes and 3 dimensions each...
INFO - Simulating Lorenz system for 1000 timesteps...
Internal standardization#
num_nodes = 3
node_dim = 3
num_timesteps = 1000
standardize = True
confounders=False,
system_name='Lorenz'
A = torch.tensor([[0,0,0],
[1,0,0],
[1,0,0]])
A, W, b, root_nodes, _ = create_scm(num_nodes,
node_dim=node_dim,
confounders=confounders,
adjacency_matrix=A)
data = simulate_system(A, W, b,
num_timesteps=num_timesteps,
standardize=standardize,
num_nodes=num_nodes,
system_name=system_name)
plot_scm(G=create_scm_graph(A), root_nodes=root_nodes)
plot_3d_trajectories(data, root_nodes, line_alpha=0.5)
plot_trajectories(data, root_nodes, sharey=False)
plt.show()
INFO - Creating SCM with 3 nodes and 3 dimensions each...
INFO - Simulating Lorenz system for 1000 timesteps...